---+ The contents below are from the old website (TBC) +---
Working Group 3 is providing guidance for the development of analytical tools for optimal positioning and functioning of the GMI platform.
GMI is bringing together scientists, public health experts, policy makers, etc. to develop a global platform (database, linked databases) that facilitates the application of NGS in research, clinical and public health settings worldwide.
Working group 3 aims to define requirements for GMI functioning from the perspective of end-users (clinical, public health, research) in terms of applications (identification, outbreak detection etc.) and priority targets/diseases. This working group wants to map current analytical options and solutions against the needs of GMI end-users, to identify possible R&D and implementation gaps and to identify projects that may fill those gaps.
- Chair: Ruth Timme, US Food and Drug Administration, Ruth.Timme@fda.hhs.gov
- Co-chairs: Heather Carleton (Centers for Disease Control and Prevention), Lee Katz (Centers for Disease Control and Prevention), and Marion Koopman (Erasmus MC)
Five sub-groups have been established that cover different aspects of WGS surveillance of microbial pathogens. Here we are including the names of these subgroups:
- WG3.1. Steering workgroup: Lead GMI WG3 annual discussions, maintain notes, email lists, summaries of accomplishments, etc.
- WG3.2 Benchmark datasets for WGS pipeline validation: Curate and expand the benchmark datasets provided on the GMI GitHub site. Maintain list of other validation efforts.
- WG3.3 Pipeline and metrics comparison: Provide guidance on which tools to use for which question on which type of dataset and run benchmarks of pipelines/tools used on GMI
- WG3.4 Metadata integration into analyses Provide tools and framework for including metadata into large-scale analyses and queries.
- WG3.5 Metagenomics: Establish standards for data sharing (sequence data and metadata); develop metagenomics benchmark datasets; develop pipeline validation guidelines
Manuscripts relevant to GMI WG3:
WGS pipeline validation for pure-isolate microbial pathogen surveillance:
- Timme RE, Strain E, Baugher J, Davis S, Gonzalez-Escalona N, Sanchez Leon M, Allard MW, Brown EW, Tallent S, Rand H 2019. Phylogenomic pipeline validation for foodborne pathogen disease surveillance. Journal of Clinical Microbiology. DOI: 10.1128/JCM.01816-18.
- Timme RE, Rand H, Shumway M, Trees EK, Simmons M, Agarwala R, Davis S, Tillman GE, Defibaugh-Chavez S, Carleton HA, Klimke WA, Katz LS 2017. Benchmark datasets for phylogenomic pipeline validation, applications for foodborne pathogen surveillance. PeerJ 5:e3893. DOI: 10.7717/peerj.3893.
- McTavish EJ, Pettengill J, Davis S, Rand H, Strain E, Allard M, Timme RE 2017. TreeToReads - a pipeline for simulating raw reads from phylogenies. BMC bioinformatics 18:178. DOI: 10.1186/s12859-017-1592-1.
- Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA 2017. A Comparative Analysis of the Lyve-SET Phylogenomics Pipeline for Genomic Epidemiology of Foodborne Pathogens. Frontiers in microbiology 8:1–13. DOI: 10.3389/fmicb.2017.00375.
- Portmann A-C, Fournier C, Gimonet J, Ngom-Bru C, Barretto C, Baert L 2018. A Validation Approach of an End-to-End Whole Genome Sequencing Workflow for Source Tracking of Listeria monocytogenes and Salmonella enterica. Frontiers in microbiology 9:446. DOI: 10.3389/fmicb.2018.00446.
- Page AJ, Alikhan N-F, Carleton HA, Seemann T, Keane JA, Katz LS 2017. Comparison of classical multi-locus sequence typing software for next-generation sequencing data. Microbial Genomics. DOI: 10.1099/mgen.0.000124.
- Kozyreva VK, Truong C-L, Greninger AL, Crandall J, Mukhopadhyay R, Chaturvedi V 2017. Validation and Implementation of Clinical Laboratory Improvements Act (CLIA)-Compliant Whole Genome Sequencing in Public Health Microbiology Laboratory. Journal of Clinical Microbiology:JCM.00361–17. DOI: 10.1128/JCM.00361-17.
- Taboada EN, Graham MR, Carriço JA, Van Domselaar G 2017. Food Safety in the Age of Next Generation Sequencing, Bioinformatics, and Open Data Access. Frontiers in microbiology 8:241. DOI: 10.3389/fmicb.2017.00909.
- Gargis AS, Kalman L, Lubin IM 2016. Assuring the Quality of Next-Generation Sequencing in Clinical Microbiology and Public Health Laboratories. Journal of Clinical Microbiology 54:2857–2865. DOI: 10.1128/JCM.00949-16.
Proficiency test exercises for pure-isolate microbial surveillance:
- Timme RE, Rand H, Sanchez Leon M, Hoffmann M, Strain E, Allard M, Roberson D, Baugher JD 2018. GenomeTrakr proficiency testing for foodborne pathogen surveillance: an exercise from 2015. Microbial Genomics 57:289. DOI: 10.1099/mgen.0.000185.
- Moran-Gilad J, Sintchenko V, Pedersen SK, Wolfgang WJ, Pettengill J, Strain E, Hendriksen RS, Global Microbial Identifier initiative’s Working Group 4 (GMI-WG4) 2015. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC infectious diseases 15:174. DOI: 10.1186/s12879-015-0902-3.
Harmonization of metadata:
- Griffiths E, Dooley D, Graham M, Van Domselaar G, Brinkman FSL, Hsiao WWL 2017. Context Is Everything: Harmonization of Critical Food Microbiology Descriptors and Metadata for Improved Food Safety and Surveillance. Frontiers in microbiology 8:1068. DOI: 10.3389/fmicb.2017.01068.
Metagenomics in Public Health Surveillance:
- Forbes Jessica D., Knox Natalie C., Ronholm Jennifer, Pagotto Franco, Reimer Aleisha 2017. Metagenomics: The Next Culture-Independent Game Changer. Frontiers in Microbiology 8:1069.
Manuscripts describing different surveillance efforts
US PulseNet/GenomeTrakr:
- Tolar B, Joseph LA, Schroeder MN, Stroika S, Ribot EM, Hise KB, Gerner-Smidt P 2019. An Overview of PulseNet USA Databases. Foodborne Pathogens and Disease:fpd.2019.2637. DOI: 10.1089/fpd.2019.2637.
- Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P 2016. Implementation of Nationwide Real-time Whole-genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 63:380–386. DOI: 10.1093/cid/ciw242.
- Timme RE, Sanchez Leon M, Allard MW 2019. Utilizing the Public GenomeTrakr Database for Foodborne Pathogen Traceback. Methods in molecular biology (Clifton, N.J.) 1918:201–212. DOI: 10.1007/978-1-4939-9000-9_17.
- Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R 2016. Practical Value of Food Pathogen Traceability through Building a Whole-Genome Sequencing Network and Database. Journal of Clinical Microbiology 54:1975–1983. DOI: 10.1128/JCM.00081-16.
Public Health England:
- Jenkins C, Dallman TJ, Grant KA 2019. Impact of whole genome sequencing on the investigation of food-borne outbreaks of Shiga toxin-producing Escherichia coli serogroup O157:H7, England, 2013 to 2017. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 24:1800346. DOI: 10.2807/1560-7917.ES.2019.24.4.1800346.
- Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, Tewolde R, Schaefer U, Jenkins C, Dallman TJ, de Pinna EM, Grant KA, Salmonella Whole Genome Sequencing Implementation Group 2016. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752. DOI: 10.7717/peerj.1752.
EU:
- Van Walle I, Björkman JT, Cormican M, Dallman T, Mossong J, Moura A, Pietzka A, Ruppitsch W, Takkinen J, European Listeria Wgs Typing Group 2018. Retrospective validation of whole genome sequencing-enhanced surveillance of listeriosis in Europe, 2010 to 2015. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 23:1073. DOI: 10.2807/1560-7917.ES.2018.23.33.1700798.
- Lüth S, Kleta S, Dahouk Al S 2018. Whole genome sequencing as a typing tool for foodborne pathogens like Listeria monocytogenes – The way towards global harmonisation and data exchange. Trends in Food Science & Technology 73:67–75.
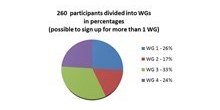
Member organisation
How members are organised in the GMI working groups.